数据来源于论文
A multi-kingdom collection of 33,804 reference genomes for the human vaginal microbiome
https://www.nature.com/articles/s41564-024-01751-5
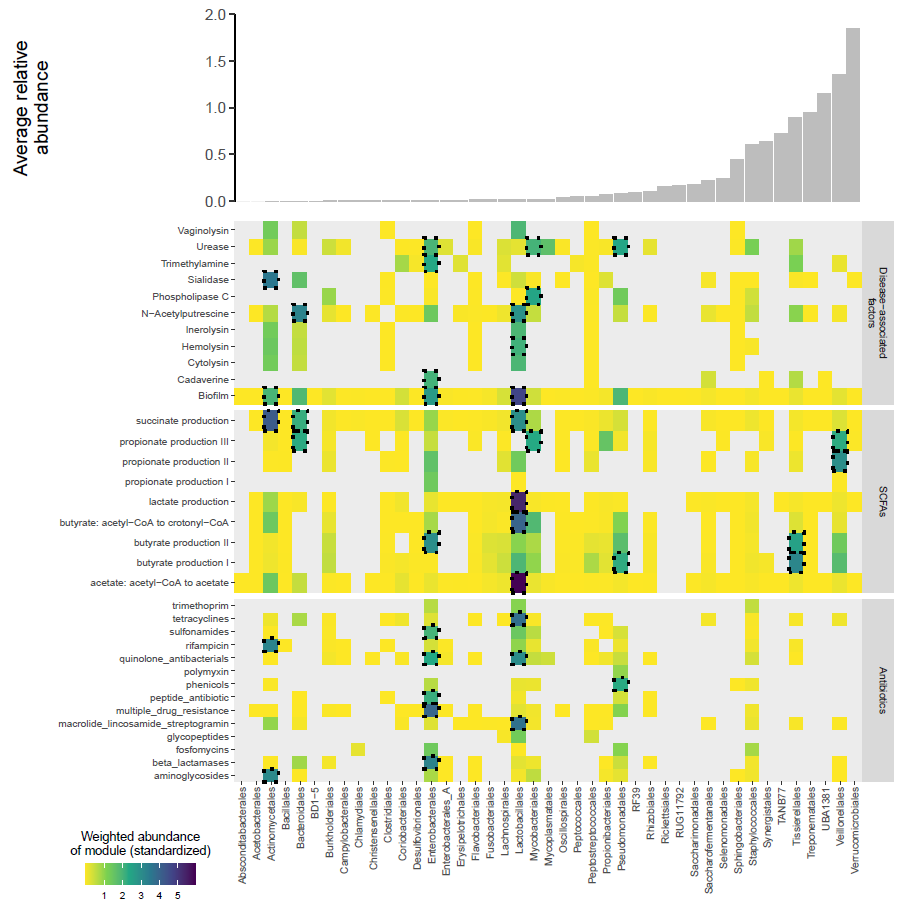
推文中复现图是论文中的figure 2e
部分示例数据截图
作图代码
library(tidyverse)
library(readxl)
library(patchwork)
fig2e.dat<-read_excel("D:/R_4_1_0_working_directory/env001/2024.data/20240717/fig2e.xlsx",
na="NA")
fig2e.dat %>% colnames()
fig2e.dat
fig2e.dat %>%
group_by(Order) %>%
summarise(mean_value=mean(`Weighted abundance of module (standardized)`,na.rm = TRUE)) %>%
ungroup() %>%
arrange(mean_value) %>%
mutate(Order=factor(Order,levels=Order))-> fig2e.bar.dat
fig2e.bar.dat %>%
pull(Order) -> Order.level
ggplot(data = fig2e.bar.dat,aes(x=Order,y=mean_value))+
geom_col(fill="#bdbdbd",color=NA)+
theme_classic(base_size = 15)+
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.line.x = element_blank())+
labs(x=NULL,y="Average relative\nabundance")+
scale_y_continuous(limits = c(0,2),
expand = expansion(mult = c(0,0))) -> fig2e.bar.plot
fig2e.bar.plot
Type.label<-c("Antibiotics\n","SCFAs\n","Disease-associated\nfactors")
names(Type.label)<-c("Antibiotics","SCFAs","Disease-associated factors")
fig2e.dat %>% pull(Type) %>%
table()
fig2e.dat %>%
mutate(Type=factor(Type,levels=c("Disease-associated factors",
"SCFAs","Antibiotics"))) -> fig2e.dat
ggplot(data = fig2e.dat,aes(x=Order,y=ID))+
geom_tile(aes(fill=`Weighted abundance of module (standardized)`))+
theme_bw(base_size = 10)+
theme(panel.border = element_blank(),
panel.grid = element_blank(),
axis.text.x = element_text(angle = 90,vjust = 0.5,hjust=1),
strip.background = element_rect(color=NA),
legend.position = c(-0.15,-0.15),
legend.direction = "horizontal",
legend.background = element_rect(fill=NA))+
scale_fill_viridis_c(direction = -1,
name="Weighted abundance\nof module (standardized)",
na.value="#ececec")+
labs(x=NULL,y=NULL)+
facet_wrap(~Type,strip.position = "right",
labeller = labeller(Type=Type.label),
ncol = 1,
scales = "free_y")+
scale_y_discrete(expand = expansion(mult = c(0,0)))+
scale_x_discrete(expand = expansion(mult = c(0,0)))+
#coord_equal()+
geom_tile(data = fig2e.dat %>%
filter(`Weighted abundance of module (standardized)`>2),
fill=NA,color="black",
lty="dotted",
linewidth=1)+
guides(fill=guide_colorbar(title.position="top",
title.hjust=0.5))+
coord_cartesian(clip = "off") -> fig2e.heatmap.plot
fig2e.heatmap.plot
fig2e.bar.plot+
fig2e.heatmap.plot+
plot_layout(ncol = 1,heights = c(1,3))
关于ggplot2的零散知识点
ggplot2分面作图不同类别的文本是strip
修改strip的文本用到的是
Type.label<-c("Antibiotics\n","SCFAs\n","Disease-associated\nfactors")
names(Type.label)<-c("Antibiotics","SCFAs","Disease-associated factors")
facet_wrap(~Type,strip.position = "right",
labeller = labeller(Type=Type.label),
ncol = 1,
scales = "free_y")
欢迎大家关注我的公众号
小明的数据分析笔记本
小明的数据分析笔记本 公众号 主要分享:1、R语言和python做数据分析和数据可视化的简单小例子;2、园艺植物相关转录组学、基因组学、群体遗传学文献阅读笔记;3、生物信息学入门学习资料及自己的学习笔记!