本期教程
获得本教程 Data and Code,请在后台回复:20240812。
2022年教程总汇
2023年教程总汇

Code
- 导入R包
library(RColorBrewer)
library(pheatmap)
library(tidyverse)
library(viridis)
library(reshape2)
filter = dplyr::filter
- 导入对应数据
input <- read.csv(file="data_Spectra_Input.csv")
head(input)
> head(input)
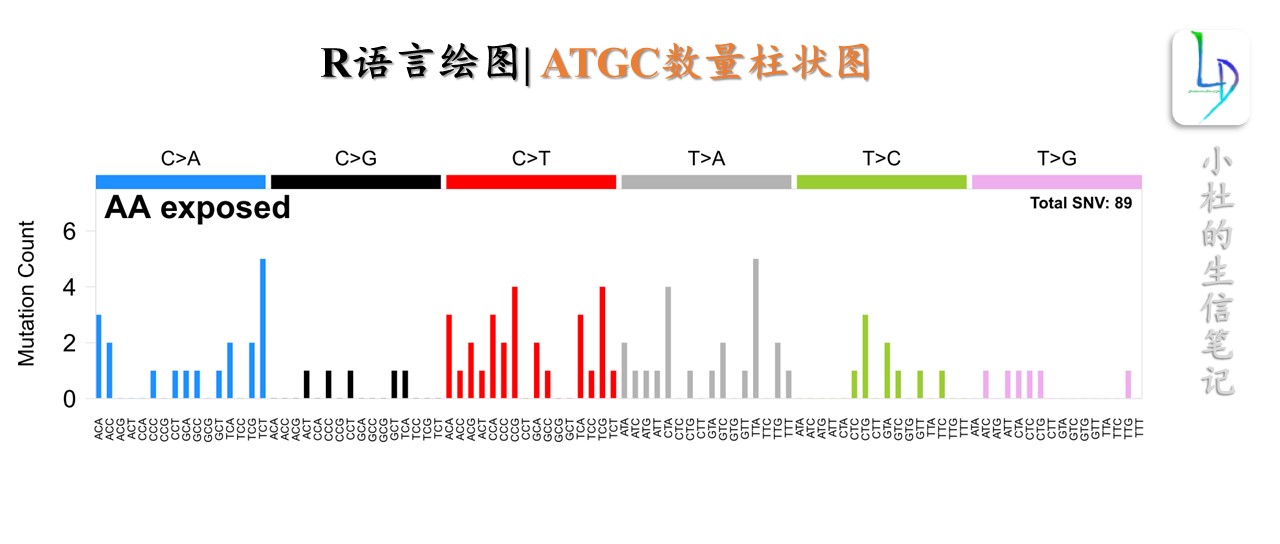
sampleID chr pos ref mut type effect gene trinuc_ref sub trinuc_ref_py firstbase thirdbase py_ref
1 PD46845a chr2 157766005 C A Sub missense ACVR1 CCT C>A CCT C T C
2 PD51536a chr14 104780108 A C Sub missense AKT1 GAG T>G CTC C C T
3 PD42673a chr14 104780214 C T Sub missense AKT1 TCC C>T TCC T C C
4 PD46732a chr5 112821985 G T Sub nonsense APC TGA C>A TCA T A C
5 PD51821a chr5 112838196 G T Sub nonsense APC AGA C>A TCT T T C
6 PD51600a chr19 46921195 C A Sub nonsense ARHGAP35 ACC C>A ACC A C C
py_alt context
1 A C[C>A]T
2 G C[T>G]C
3 T T[C>T]C
4 A T[C>A]A
5 A T[C>A]T
6 A A[C>A]C
AA_exposed <- read.csv(file="data_AA_Input.csv")
head(AA_exposed)
> head(AA_exposed)
Sample Country SBS22a SBS22b SBS22a_relative SBS22b_relative
1 PD37463a Romania 6878 8024 0.3119 0.3639
2 PD37464a Romania 660 1049 0.1196 0.1901
3 PD37466a Romania 1349 1301 0.2088 0.2014
4 PD37469a Romania 0 2539 0.0000 0.3993
5 PD37484a Romania 3718 7772 0.2712 0.5670
6 PD42572a Serbia 4015 3463 0.3788 0.3268
- 提取、计算、分析
### 绘图函数
plot_driver_spectra = function(input, plot_title) {
#Collect Data
mutations = input
#Define contexts
matrix_contexts <- c('A[C>A]A', 'A[C>A]C', 'A[C>A]G', 'A[C>A]T', 'C[C>A]A', 'C[C>A]C', 'C[C>A]G', 'C[C>A]T',
'G[C>A]A', 'G[C>A]C', 'G[C>A]G', 'G[C>A]T', 'T[C>A]A', 'T[C>A]C', 'T[C>A]G', 'T[C>A]T',
'A[C>G]A', 'A[C>G]C', 'A[C>G]G', 'A[C>G]T', 'C[C>G]A', 'C[C>G]C', 'C[C>G]G', 'C[C>G]T',
'G[C>G]A', 'G[C>G]C', 'G[C>G]G', 'G[C>G]T', 'T[C>G]A', 'T[C>G]C', 'T[C>G]G', 'T[C>G]T',
'A[C>T]A', 'A[C>T]C', 'A[C>T]G', 'A[C>T]T', 'C[C>T]A', 'C[C>T]C', 'C[C>T]G', 'C[C>T]T',
'G[C>T]A', 'G[C>T]C', 'G[C>T]G', 'G[C>T]T', 'T[C>T]A', 'T[C>T]C', 'T[C>T]G', 'T[C>T]T',
'A[T>A]A', 'A[T>A]C', 'A[T>A]G', 'A[T>A]T', 'C[T>A]A', 'C[T>A]C', 'C[T>A]G', 'C[T>A]T',
'G[T>A]A', 'G[T>A]C', 'G[T>A]G', 'G[T>A]T', 'T[T>A]A', 'T[T>A]C', 'T[T>A]G', 'T[T>A]T',
'A[T>C]A', 'A[T>C]C', 'A[T>C]G', 'A[T>C]T', 'C[T>C]A', 'C[T>C]C', 'C[T>C]G', 'C[T>C]T',
'G[T>C]A', 'G[T>C]C', 'G[T>C]G', 'G[T>C]T', 'T[T>C]A', 'T[T>C]C', 'T[T>C]G', 'T[T>C]T',
'A[T>G]A', 'A[T>G]C', 'A[T>G]G', 'A[T>G]T', 'C[T>G]A', 'C[T>G]C', 'C[T>G]G', 'C[T>G]T',
'G[T>G]A', 'G[T>G]C', 'G[T>G]G', 'G[T>G]T', 'T[T>G]A', 'T[T>G]C', 'T[T>G]G', 'T[T>G]T')
#Count Occurances
freq=c(table(mutations$context))
mutations_ctx=data.frame(ctxt=matrix_contexts,counts=freq[matrix_contexts])
mutations_ctx <- tibble::column_to_rownames(mutations_ctx,"ctxt")
mutations_ctx[is.na(mutations_ctx)] = 0
rownames(mutations_ctx) <- NULL
#Plot!
# Specify Context Type
sig_cat = c("C>A","C>G","C>T","T>A","T>C","T>G")
ctx_vec = paste(rep(c("A","C","G","T"),each=4),rep(c("A","C","G","T"),times=4),sep="-")
full_vec = paste(rep(sig_cat,each=16),rep(ctx_vec,times=6),sep=",")
snv_context = paste(substr(full_vec,5,5), substr(full_vec,1,1), substr(full_vec,7,7), sep="")
# Specify Vectors for plot colours
col_vec_num <- rep(16,6)
col_vec = rep(c("dodgerblue","black","red","grey70","olivedrab3","plum2"),each=16)
# Convert to matrix
sig_plot <- as.matrix(mutations_ctx)
# Set up Signature Names and title
sig_title <- colnames(sig_plot)
# Set up Counts
sample_counts <- colSums(mutations_ctx)
# Set par settings
par(xaxs='i', cex = 1, xpd = FALSE)
# define maxy
max_prob <- sig_plot[,1];maxy = max(max_prob)
# call empty plot
b = barplot(sig_plot[,1], col = NA, border = NA, axes = FALSE, las = 2, ylim=c(0,1.5*maxy),
names.arg = snv_context, cex.lab = 1.3, cex.names = 0.70, cex.axis = 2,
space = 1, ylab = "Mutation Count")
# add axis
axis(2, at = pretty(0:(1.5*maxy), n = 3), col = 'grey90', las = 1, cex.axis = 1.5)
# call columns
b = barplot(sig_plot[,1], axes = FALSE, col=col_vec, add = T, border = NA, space = 1)
# add box surronding plot
box(lty = 1, col = 'grey90')
# add title
title(plot_title, line = -1.5, adj = 0.01, cex.main = 2)
title(paste0("Total SNV: ", sample_counts), line = -1, adj = 0.99, cex.main = 1)
# add rectangles and annotations on top of the plot
par(xpd = NA)
for (j in 1:length(sig_cat)) {
xpos = b[c(sum(col_vec_num[1:j])-col_vec_num[j]+1,sum(col_vec_num[1:j]))]
rect(xpos[1]-0.5, maxy*1.5, xpos[2]+0.5, maxy*1.6, border=NA, col=unique(col_vec)[j])
text(x=mean(xpos), pos=3, y=maxy*1.6, label=sig_cat[j], cex = 1.25)
}
}
- 提取、计算、分析
#Subset only for cases with >10% COSMIC attribution to either AA signatures
input_AA <- subset(input, (sampleID %in% AA_exposed$Sample))
input_nonAA <-subset(input, (!sampleID %in% AA_exposed$Sample))
- 绘图
plot_driver_spectra(input_AA,"AA exposed")

获得本教程 Data and Code,请在后台回复:20240812。
若我们的教程对你有所帮助,请 点赞+收藏+转发,这是对我们最大的支持。
往期部分文章
1. 最全WGCNA教程(替换数据即可出全部结果与图形)
2. 精美图形绘制教程
3. 转录组分析教程
4. 转录组下游分析
小杜的生信筆記 ,主要发表或收录生物信息学教程,以及基于R分析和可视化(包括数据分析,图形绘制等);分享感兴趣的文献和学习资料!!