论文
Genomic insights into local adaptation and future climate-induced vulnerability of a keystone forest tree in East Asia
https://www.nature.com/articles/s41467-022-34206-8
论文中基本提供了每个图的原始数据,也提供了大部分代码,原始数据在电脑上的存储名 41467_2022_34206_MOESM8_ESM.zip
论文中提供的代码链接
https://github.com/jingwanglab/Populus_genomic_prediction_climate_vulnerability
但是在这部分代码里没有找到 做mantel的代码
mantel检验主要是用来做两个距离矩阵之间的相关性
论文中关于这部分分析的方法部分
To investigate and compare the role of geography and environment in shaping spatial genetic variation of adaptive (the 1779 adaptive variants identified by both LFMM and RDA) and neutral (the 535,191 LD-pruned SNPs as used for population structure analyses) variants, Mantel and partial Mantel tests were separately used to test for associations between _F_ST(_F_ST/1−_F_ST) and geographic (IBD) and environmental (IBE) distance (after accounting for the geographic distance) with significance determined using 999 permutations in the R package vegan87, where environmental distance was represented by Euclidean distance of all scaled environmental variables.
分子距离这里用的是FST相关,但是为什么用 _F_ST(_F_ST/1−_F_ST) 这个值暂时没有想明白。地理距离的算法在 Genetic subdivision and candidate genes under selection
in North American grey wolves 这篇论文里有提到
https://datadryad.org/stash/dataset/doi:10.5061/dryad.c9b25
地理距离是用Genalex 这个软件算的
这篇论文里提到的分子距离是用plink算的
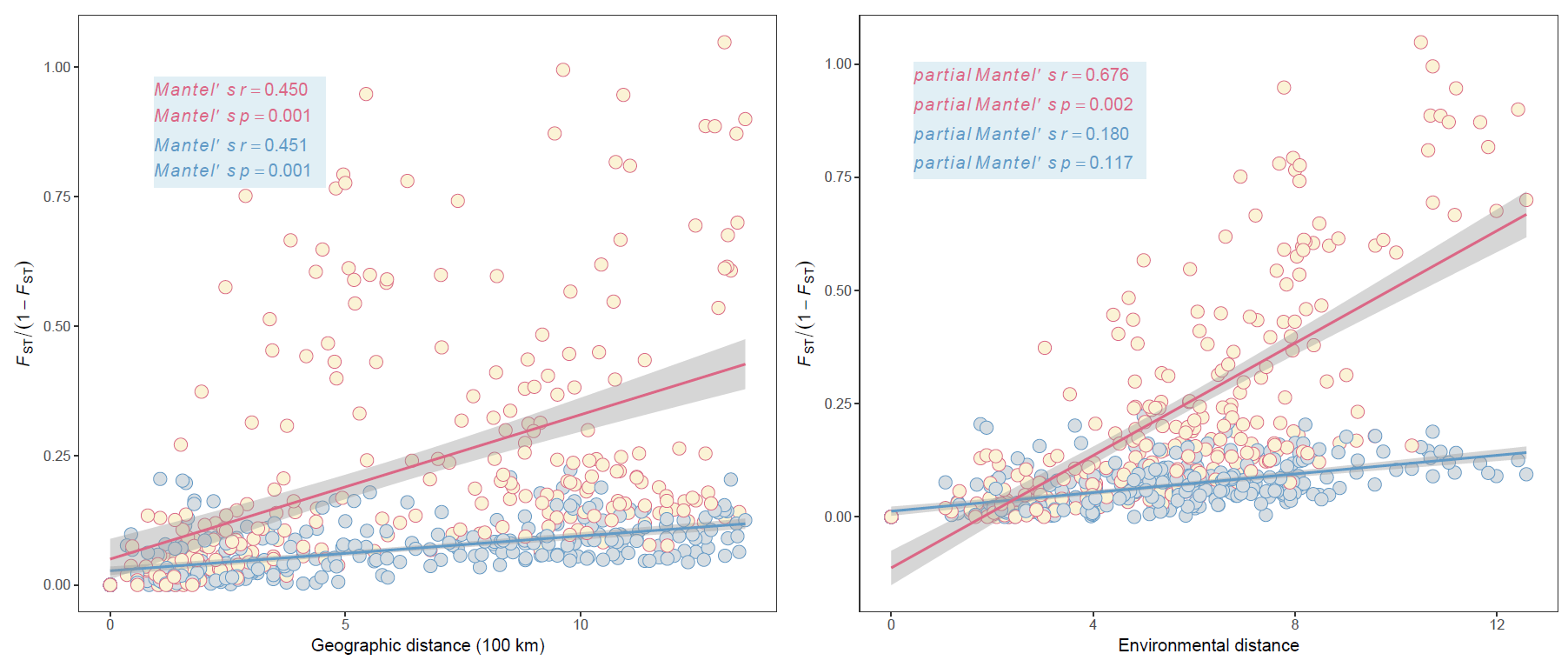
NC的论文里已经提供了算好的距离矩阵,今天的推文里就直接用距离矩阵做mantel检验,复现论文中的Fig2C
读取数据
library(tidyverse)
read.csv("data/20221211/Sourcedata/Fig2c&d/Fst_adaption.csv",
row.names = 1) %>%
as.matrix() %>%
Matrix::tril() %>%
as.dist() -> fst.adaption.dist
read.csv("data/20221211/Sourcedata/Fig2c&d/Fst_neutral.csv",
row.names = 1) %>%
as.matrix() %>%
Matrix::tril() %>%
as.dist() -> fst.neutral.dist
read.csv("data/20221211/Sourcedata/Fig2c&d/geo_dist.csv",
row.names = 1) %>%
as.matrix() %>%
Matrix::tril() %>%
as.dist() -> geo.dist
这里提供的数据是一个对称矩阵,读取进来以后是一个数据框,最后转换成了一个下三角矩阵用于 mantel函数的输入
做mantel检验的函数
vegan::mantel(fst.adaption.dist,geo.dist,permutations = 999)
vegan::mantel(fst.neutral.dist,geo.dist,permutations = 999)

输出结果和论文中的一致
做partial mantel检验
vegan::mantel.partial(fst.adaption.dist,env.dist,geo.dist,permutations = 999)
vegan::mantel.partial(fst.neutral.dist,env.dist,geo.dist,permutations = 999)
这个结果和论文中的也一致
作图代码
dat<-read_csv("data/20221211/Sourcedata/Fig2c&d/plot_data.csv")
tt1<-ttheme_minimal(core=list(fg_params=list(hjust=0,
parse=TRUE,
x=0,
fontsize=15,
col=c("#db6786","#db6786",
"#609ac6","#609ac6")),
bg_params=list(fill="#e1eff5")))
table2c<-tibble(x=c("italic(Mantel)*minute*italic(s)~italic(r)=='0.450'",
"italic(Mantel)*minute*italic(s)~italic(p)=='0.001'",
"italic(Mantel)*minute*italic(s)~italic(r)=='0.451'",
"italic(Mantel)*minute*italic(s)~italic(p)=='0.001'"))
p2c<-dat %>%
select(geo_dit,fst1779,fst53w) %>%
pivot_longer(!geo_dit) %>%
ggplot(aes(x=geo_dit/100000,y=value))+
geom_point(aes(color=name,
fill=name),
shape=21,
size=5)+
scale_color_manual(values = c("#609ac6","#db6786"))+
scale_fill_manual(values = c("#d6dde2","#fbf3d5"))+
geom_smooth(aes(color=name),
method = "lm",
formula = 'y~x')+
theme_bw(base_size = 15)+
theme(panel.grid = element_blank(),
legend.position = "none")+
labs(x="Geographic distance (100 km)",
y=expression(italic(F)[ST]/(1-italic(F)[ST])))+
annotation_custom(tableGrob(table2c,
rows = NULL,
cols = NULL,
theme = tt1),
xmin=0.5, xmax=5, ymin=0.75, ymax=1)
p2c
table2d<-tibble(x=c("italic(partial~Mantel)*minute*italic(s)~italic(r)=='0.676'",
"italic(partial~Mantel)*minute*italic(s)~italic(p)=='0.002'",
"italic(partial~Mantel)*minute*italic(s)~italic(r)=='0.180'",
"italic(partial~Mantel)*minute*italic(s)~italic(p)=='0.117'"))
p2d<-dat %>%
select(env_dist,fst1779,fst53w) %>%
pivot_longer(!env_dist) %>%
ggplot(aes(x=env_dist,y=value))+
geom_point(aes(color=name,
fill=name),
shape=21,
size=5)+
scale_color_manual(values = c("#609ac6","#db6786"))+
scale_fill_manual(values = c("#d6dde2","#fbf3d5"))+
geom_smooth(aes(color=name),
method = "lm",
formula = 'y~x')+
theme_bw(base_size = 15)+
theme(panel.grid = element_blank(),
legend.position = "none")+
labs(x="Environmental distance",
y=expression(italic(F)[ST]/(1-italic(F)[ST])))+
annotation_custom(tableGrob(table2d,
rows = NULL,
cols = NULL,
theme = tt1),
xmin=0.5, xmax=5, ymin=0.75, ymax=1)
p2d
library(patchwork)
p2c+p2d
欢迎大家关注我的公众号
小明的数据分析笔记本
小明的数据分析笔记本 公众号 主要分享:1、R语言和python做数据分析和数据可视化的简单小例子;2、园艺植物相关转录组学、基因组学、群体遗传学文献阅读笔记;3、生物信息学入门学习资料及自己的学习笔记!